Baseball Activity and
Player Recognition from Inertial Sensor Data
Gershon Dublon and Jared Markowitz
MAS 622J/1.126J
Pattern Recognition and Analysis
Final Project Report
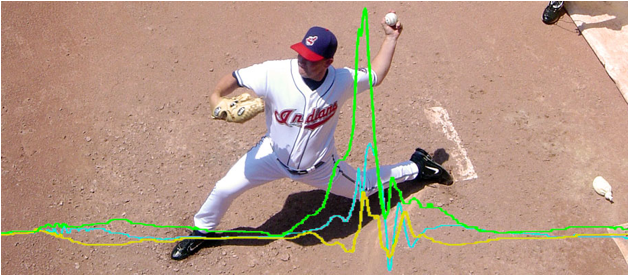
Introduction and
Motivation
Baseball
is a sport known for the extensive statistical analyses teams employ to gain an
edge. Major League organizations
spend huge amounts of time and money evaluating player performance through a
variety of statistical methods.
Similarly they dedicate many resources toward keeping their players
healthy, doing everything they can to enable peak performance. To this point there has been relatively
little effort to merge the worlds of statistical analysis and sports medicine;
in particular there has been relatively little effort expended to quantify the
mechanics of baseball movements at a sufficiently high resolution to understand
the enormous forces and torques the game exerts on the joints of its players.
These quantities are of particular interest to sports medicine researchers who
seek to better understand the common causes of injury, catch problematic
behaviors before they cause long-term damage, and design safer practice and
play guidelines for professional athletes. Beyond sports medicine, researchers
are beginning to evaluate quantitative metrics to characterize player
performance and skill in order to design of more effective and targeted
training regimens.

Motion
capture systems are the quantitative tools most commonly applied to this
problem, but fail to capture the intricacies of player technique. This results from their lack of
spatiotemporal resolution and the precise setup and calibration they require,
which effectively precludes portability to the ballfield. This project
leverages sportSemble, a sensing
platform built by Lapinksi et al, of the Responsive
Environments Group at the MIT Media Lab [1]. sportSemble
consists of a set of small, high-precision, high-range inertial measurement
units (IMUs) designed for sports medicine and performance evaluation. The sportSemble system enables high-resolution measurements of
the linear accelerations and angular velocities of the relevant limbs and
joints, while avoiding the pitfalls of the traditional motion-capture approach.
In
this project, we take a first pass at a dataset of professional pitchers and
batters, aiming to extract and map a feature space of these data and discover
good features for classification of a number of player activities: pitch type, swing
type, and the resultant ball locations for pitches and swings. We also report
on feature selection and classification of player identification, using the
same motion signatures as biometric identifiers. It is worth noting that these
problem differ significantly (in data and feature-space) from the classical
IMU-centric “activities of daily living” (ADL) recognition problem because the
relevant examples are all of duration less than one second and involve
accelerations, angular velocities, and fine motor control well beyond most
human capability. We select and test features, reporting the results for a
number of classification schemes, including naēve Bayes, k-Nearest Neighbor,
Linear Discriminants, and Support Vector Machines.
Dataset
The
sportSemble dataset is collected from professional
baseball players who are directed to perform common tasks (pitching and
swinging). The players wear five 6-degree-of-freedom (6-DOF) IMUs on their
hands, forearms, upper arms, chests, and waists. Each IMU consists of two
3-axis accelerometers and two 3-axis gyroscopes (one of each for high- and
low-range measurements), as well as a 3-axis magnetometer (which we did not
consider) and a radio for synchronization. The data is stored to local flash on
each IMU at a sample rate of 1000 Hz for the high-range and 330 Hz for the
low-range sensors. Each example includes 8 seconds of data for each axis of
each sensor, adding up to slightly under 300,000 data points per example. The labels are annotated during data
collection and manually entered into a database (together with the data) later.
These data and labels are retrieved from the database as needed.
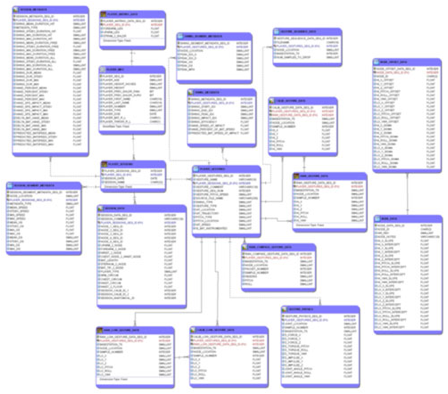
Figure
1: Learning PostgreSQL: the structure of the sportSemble database
For
the purposes of classification, there were insufficient numbers of examples for
several of the relevant classes.
For example, there were only 3 curveballs in the entire set. Since very
little can be extrapolated from the correct (or incorrect) classification of
such small numbers of examples, we discarded those out of hand (though in
clustering and plotting we did find that 2 of the 3 curve ball examples were
very close together and extremely far in 3-feature space from any other
examples). Data corresponding to unrealistically small classes was retained for
the other classification problems if its other labels were sufficient (i.e.
curve balls cannot be classified, but their locations can). After a tedious
process of weeding out bad (glitchy, missing,
unacceptably noisy) data and examples in which the relevant activity is
partially cut off in time (either at the start or end of the data), we were
left with the following numbers of each motion. The number of examples per
class are given below to convey a sense of the baseline (naēve) prior
performance we hoped to beat.
|
Pitch
Type |
No.
Examples |
|
Pitch
Location* |
No.
Examples |
|
|
Fastball |
131 |
|
Left |
78 |
|
|
Change |
39 |
|
Middle |
114 |
|
|
Slider |
9 |
|
Right |
41 |
|
|
2-seam |
17 |
|
Top |
18 |
|
|
4-seam |
7 |
|
Middle |
139 |
|
|
Break |
27 |
|
Bottom |
76 |
|
|
Total |
230 |
|
Total |
233 |
|
|
|
|
|
|||
|
Swing
Type |
No.
Examples |
|
Swing
Location* |
No.
Examples |
|
Tee |
91 |
|
Left |
60 |
|
Soft Toss |
90 |
|
Center |
76 |
|
Normal |
33 |
|
Right |
78 |
|
Total |
214 |
|
Total |
214 |
Table
1: Motion types and number of each that were used in this analysis.
|
Player
ID (pitchers) |
No.
Examples |
|
Player
ID (batters) |
No.
Examples |
|
#2 |
22 |
|
#1 |
15 |
|
#3 |
33 |
|
#12 |
20 |
|
#4 |
33 |
|
#13 |
12 |
|
#5 |
36 |
|
#15 |
19 |
|
#6 |
42 |
|
#16 |
17 |
|
#8 |
24 |
|
#18 |
19 |
|
#9 |
21 |
|
#21 |
44 |
|
#31 |
22 |
|
#23 |
14 |
|
Total |
233 |
|
#24 |
15 |
|
|
|
|
#25 |
18 |
|
|
|
|
#27 |
21 |
|
|
|
|
Total |
214 |
* Location
labels were flipped for left-handed pitchers and batters so as to be symmetric
with respect to the IMU axes on the right-handed players.
Table
2: Player ID labels and numbers of each for pitching and swinging
Procedure
1.) From the
raw data described above, we extracted 196 pitching features and 200 batting
features.
2.) Several
methods of feature selection were considered to determine the features relevant
to several different classification problems. Sequential Forward Selection was used to
determine final features.
3.) The
performances of four classification schemes (NB, kNN,
LDA, SVM) were evaluated and used to improve understanding of the data set.
1.
Feature Extraction
Extracting
good features from the data was a challenge because there is a large amount
data per example. It is not immediately obvious which data is most relevant for
the extraction of salient features, and what the most salient features are. For
pitchers, we used the high-range (higher sampling rate, lower bit depth) sensor
data for the hand, forearm, and upper arm, and low-range (lower sampling rate,
higher bit depth) for the waist and chest. For batters, we used the high-range
sensor data for the hand and forearm, and low-range for the upper arm, waist
and chest. These choices reflect the magnitude of the motion recorded at each
IMU, which saturates the low-range sensors on all the players’ hands and
forearms, and on pitchers’ upper arms.
i. Peak timing
Given
the peakiness of the raw data and relatively short time spans involved, we
hypothesized that relative peak timing between axes and across IMUs would be
most salient for our classification problems. We encoded this feature by
defining an “origin” for each pitching example to be the highest peak in the
z-axis of the hand (pointing out in the direction of the hand), consistently
the highest and sharpest peak. The origin for each batting example was given by
the impact time, a pre-computed feature by Lapinski,
et al. Once the origin sample index was extracted, we extracted the 2 highest
peaks from each remaining IMU (accelerometer and gyroscope) axis and retained
the time between those peaks as features.
ii.
Relative peak heights and inertial magnitudes
We
extracted the heights of the 2 highest peaks of the rectified signal in each
axis and from the IMU magnitudes of acceleration and rotation. We then
“normalized” these values by the height of the highest peak in the data (hand
acceleration in the z-axis) in an attempt to make the features more invariant
to overall player strength. These relative peak heights were recorded as
features.
iii.
Spectral peaks
We
computed a zero-padded, 1024-point Fast Fourier Transform (FFT) for each axis,
and retained the frequencies corresponding to 3 peaks in the spectrum for those
signals that had periodic characteristics. For example, the hand acceleration
in z had no peaks in the spectrum (as it is an impulse in time), while the x
and y axes in that IMU tended to have clear spectral peaks.
iv.
Full-width at half maximum (FWHM)
We
computed the full duration at half-maximum of the highest peaks of the
magnitudes of acceleration and rotation in each IMU, intended to encode the
sharpness of the peaks.
v. Impact
timing
We
used pre-computed start, impact, and end times provided by Lapinksi,
et al. for the batting data, stored relative to impact time. The resulting two
features encode the point in the swing when the ball is hit, which is presumed
relevant for ball location.
A
table of contents of features for pitching is linked here:
A
table of contents of features for batting is linked here:
Feature
Selection
The
above methods of feature extraction produced very large number of features for
each movement, relative to the number of example, almost one-to-one. We tried a
number of methods to avoid overfitting and other pitfalls of huge feature
spaces.
i. Principal Components Analysis
First,
to get a sense of the variance in the feature space, we computed its principal
components. Principle Component Analysis (PCA) uses an orthogonal
transformation to produce linear combinations of the original features that are
uncorrelated, with the resulting components being listed in order of decreasing
variance. This analysis exposes the correlation between features, and shows
where the variance of the feature space is concentrated.
Results
of this step are shown in Figure 2 which shows that the dominant components
best separate individual players (as opposed to classes), indicating that the
features of highest variance vary significantly more across individuals than
across motion classes. Indeed, only 2 components account for 93% of the
variance in the set. These results were initially discouraging, though they
suggested immediately that certain features that are especially good for
separating players would not be good, and that variance in the feature space is
not necessarily a good indicator of goodness.
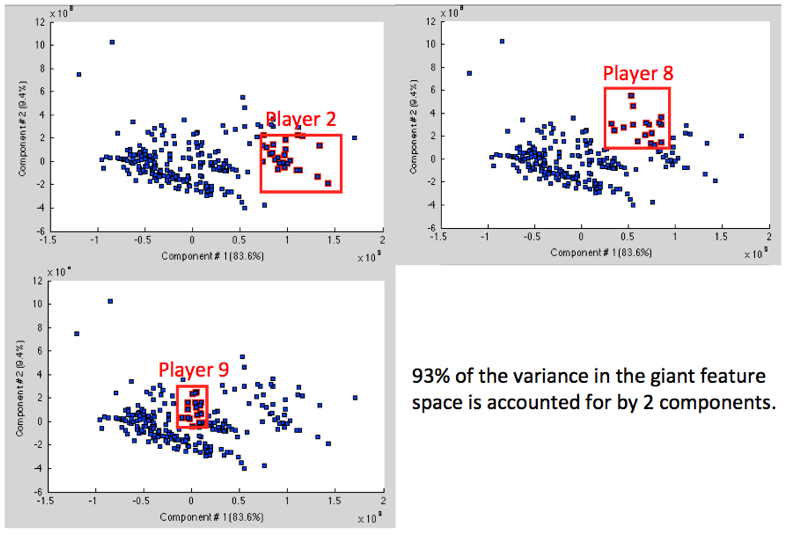
Figure
2: First two components of PCA account for almost all the variance in the
feature space, and separate players into clusters
ii. Mutual
Information
Several
techniques were then applied to determine which individual features are most
closely linked with labels. The
labels we refer to here are pitch type and pitch location, hit location, and
player ID. To start, we first
computed the mutual information, I(X; Y)
of each feature X with each output
label Y.
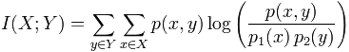
Mutual
information measures how much the distributions of the variables differ from
statistical independence, and thus here measures the dependence of the output
label with each individual feature.
An example plot of how mutual information varies across the features is
shown below. While mutual
information does give a sense of how much the output is correlated with each
input, it does not consider potential interaction between features in producing
an output and hence is not an ideal feature selector. Still, the
distinguishable peaks in this quantity indicate features that are correlated
with the labels. Figure 3 shows more variation and peakiness in the mutual
information for pitch type than hit location.
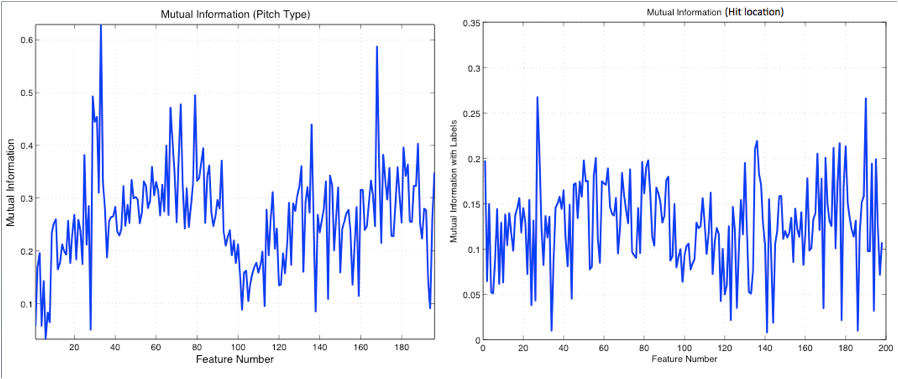
Figure 3: Mutual Information
vs. Feature Number for pitch type and hit location
iii.
Sequential Forward Selection
To
better understand how the interactions between features affect the output
labels, we proceeded to apply a Sequential Forward Selection (SFS) algorithm to
select features. Such an algorithm
is based on the accuracy of an individual classification method on
predetermined subsets of the data (training and testing sets). The algorithm proceeds in a “greedy”
manner, starting with no features and then keeping the feature that most
improves the classification accuracy in each run through the data. The algorithm can be terminated at
either a predetermined number of features or once the addition of another
feature yields only a small increase in performance. In this study SFS was applied with 3
different classifiers (NB, kNN, LDA) with varying
success, as described below. The
classification performance was evaluated using both the leave-one-out and
10-fold cross-validation methods for dividing the data into training and
testing sets. The leave-one-out
method selects a single example for classification, using the rest of the data set
for training. This process is repeated for all points in the set, and an
over-all accuracy is then computed. The 10-fold cross-validation method
randomly partitions the data set into 10 subsets and then, for each subset,
classifies the points using the other 9 subsets as training data. The overall accuracy of this method is
then the classification accuracy over all 10 subsets. The performance of the
10-fold cross-validation is likely to be more realistic (or is less likely to overfit) than leave-one-out as it relies on less training
data for each classification and does not have an individual training set for
each point. Below we discuss the
results of this classification scheme for each of the different classification
problems and classifiers that were pursued in this study.
Classifiers
i. k-Nearest Neighbor (k-NN)
In
this classifier, a given point is classified according to the classes of the k
training samples nearest it in the feature space (here the Euclidean distance
is used). The point in question is
sorted into the class that contains the largest number of the k surrounding
points. The kNN algorithm works well on data sets
where the classes are distributed in largely homogeneous clusters (including
those with multiple clusters per class).
ii. Naive
Bayes
The
Naive Bayes classifier assumes that the value of a given feature is
conditionally independent of that of any other feature, given the class. The
classifier then chooses the class with the highest conditional probability
given the feature observations, based on the training set.
iii. Linear
Discriminants (LDA)
Linear
discriminant classification learns the linear combination of features that best
separates the classes in the feature space. In general, linear classifiers work
best separating classes that are not distributed in clusters throughout the
feature space, but easily separated by hyperplanes. LDA performance can often
be improved by combining weak-performing linear classifiers through Adaptive
Boosting or other meta-algorithms.
iv. Support
Vector Machines (SVM)
At
the most basic level, SVM classifiers find a separating hyperplane in the
feature space, similar to the LDA classifier. In contrast to LDA, SVMs can map
the feature space to a higher dimensional one, such that classes that are not
linearly separable in the original feature space become separable in the new
one. In particular, SVMs with radial basis function kernels are well suited to
feature spaces where a single class is divided in the space by another, because
they bend the space along the line joining the separated class clusters to join
the class before applying the separating hyperplane. This principle is further
elucidated below.
out
Results
and Discussion
The
results of the Sequential Forward Selection (SFS) algorithms used here are
displayed in Tables 3, 4 and 5. Six
different classification problems were analyzed: pitch type (fastball /
changeup / slider / two-seam fastball / four-seam fastball / breaking ball),
pitch horizontal location (left / right / center), pitch vertical location (up
/ center / down), pitcher identification, hit direction (left / center /
right), and batter identification.
Table 3 and Table 4 show the results using leave-one-out and 10-fold
cross-validation for choosing training and testing sets. It should be noted that the selection
applying leave-one-out cross-validation always tried up to 20 features and
picked the number with the best classification accuracy, while the selection
using 10-fold validation stopped as soon as the addition of one feature failed
to improve classification accuracy.
Five different values of k in the k-NN algorithm were tested in each
case, with the best k being reported in the tables. As shown in Figure 4 (A), the
classification accuracies in most problems did not change appreciably for k
varying from 1 to 11 (with the exception of the batter identification
problem). Figure 4 (B) shows the convergence
of the errors in the player ID problem using the LDA and k-NN classifiers.
Leave-one-out
Cross-Validation
|
Class. Problem |
Method |
# Features |
Top 10 (or less) Features |
Error |
|
Pitch Type |
kNN5 |
4 |
29,79,33,66 |
0.1803 |
|
Pitch Type |
LDA |
1 |
141 |
0.5107 |
|
Pitch Horiz. |
kNN9 |
8 |
165,34,109,159,67,123,56,126 |
0.382 |
|
Pitch Horiz. |
LDA |
4 |
43,4,138,29 |
0.4721 |
|
Pitch Vert. |
kNN3 |
2 |
148,85 |
0.3777 |
|
Pitch Vert. |
LDA |
1 |
138 |
0.4421 |
|
Pitch Player ID |
kNN1 |
8 |
68, 153,96,31,24,60,97 |
0.0043 |
|
Pitch Player ID |
LDA |
4 |
61,174,180,53 |
0.0086 |
|
Hit Direction |
kNN3 |
4 |
50,143,62,128 |
0.5047 |
|
Hit Direction |
LDA |
4 |
28,84,60,6 |
0.5093 |
|
Hit Direction |
NB |
3 |
179,138,121 |
0.5561 |
|
Hit Player ID |
kNN1 |
8 |
79,57,93,89,55,125,134,128 |
0.0467 |
|
Hit Player ID |
LDA |
9 |
57,89,59,79,173,189,82,188,61 |
0.0187 |
Table 3 : SFS results using
leave-one-out cross-validation and kNN, LDA, NB
classifiers.
10-fold Cross-Validation
|
Class. Problem |
Method |
# Features |
Top 10 (or less) Features |
Error |
|
Pitch Type |
kNN1 |
7 |
29,33,58,65,67,79,89 |
0.1783 |
|
Pitch Type |
LDA |
6 |
7,29,115,141,142,167 |
0.4739 |
|
Pitch Horiz. Loc. |
kNN1 |
4 |
8,71,84,105 |
0.4378 |
|
Pitch Horiz. Loc. |
LDA |
2 |
43,138 |
0.4893 |
|
Pitch Horiz. Loc. |
NB |
3 |
77,130,196 |
0.4635 |
|
Pitch Vert. Loc. |
kNN7 |
3 |
57,82,153 |
0.3348 |
|
Pitch Vert. Loc. |
LDA |
1 |
138 |
0.4549 |
|
Pitch Vert. Loc. |
NB |
3 |
68,77,128 |
0.3605 |
|
Pitch Player ID |
kNN1 |
8 |
55,57,65,66,67,126,144 |
0.0701 |
|
Pitch Player ID |
LDA |
5 |
47,61,136,174,180 |
0.0093 |
|
Hit Location |
kNN1 |
5 |
27,46,53,96,125 |
0.4673 |
|
Hit Location |
LDA |
2 |
28,84 |
0.5234 |
|
Hit Location |
NB |
2 |
89,95 |
0.5794 |
|
Hit Player ID |
kNN5 |
8 |
55,57,59,79,89,91,93,167 |
0.0701 |
|
Hit Player ID |
LDA |
16 |
15,17,51,53,57,58,64,73,87,122 |
0.0093 |
Table 4: SFS results for best kNN, LDA, and NB using 10-fold cross-validation.
|
Class. Problem |
Method |
# Features |
Top 10 (or less) Features |
Error |
|
Pitch Type- FAST |
SVM |
2 |
141,168 |
0.2696 |
|
Pitch Type-CHANGE |
SVM |
2 |
27,184 |
0.1391 |
|
Pitch Horiz. Loc. - L |
SVM |
2 |
27,82 |
0.2935 |
|
Pitch Horiz. Loc. – C |
SVM |
2 |
4,63 |
0.3312 |
|
Pitch Horiz. Loc. – R |
SVM |
3 |
63,84,115 |
0.1438 |
|
Pitch Vert. Loc. –HI |
SVM |
2 |
105,136 |
0.0594 |
|
Pitch Vert. Loc.- MI |
SVM |
4 |
58,104,139,146 |
0.3215 |
|
Pitch Vert. Loc.- LO |
SVM |
5 |
56,92,108,120,154 |
0.2489 |
|
Hit Location – L |
SVM |
1 |
103 |
0.3076 |
|
Hit Location – C |
SVM |
4 |
16, 58,122,146 |
0.2394 |
|
Hit Location - R |
SVM |
3 |
131,136,148 |
0.3112 |
Table 5: SFS results for SVM
classifier using 10-fold cross-validation. Results are omitted for the smallest
classes due to overfitting.
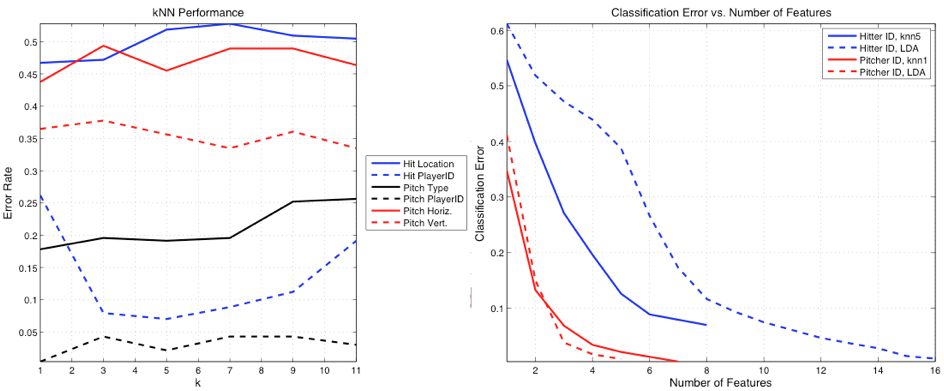
Figure
4: (A) k-NN performance versus k.
(B) Reduction of error with number of features in SFS.
Several
features of tables 3,4 and 5 stand out.
The majority of the features chosen by SFS relate to the timing of the
accelerometer and gyroscope data on the hand and forearms, confirming
expectations given the motions recorded.
The exceptions to this trend are the player ID classification problems,
where measurements taken at the waist and upper arm are found to be more
important. One explanation for this would be that the differing body structures
and strengths of the different players could be used to identify them and would
be better reflected in the data from these proximal sensors. Overall, many different features were
found to be relevant depending on the problem and the approach. Given the redundancies in our “kitchen
sink” approach to feature extraction, this is not an unexpected result;
however, the small size of the dataset relative to the feature space
significantly increases the risk of overfitting. We clearly saw these effects
on the smallest classes, and especially given the binary training and
classification of SVM. For example, SVMs achieved a 1% error rate after feature
selection on the pitch type slider (with only 9 examples in the set), and 27%
error for fastballs (which account for half the dataset). As a result, we omit
from this report the results of feature selection using SVM for the smaller
classes, which can only be accounted for by overfitting. On the bright side,
the feature selection framework is in now in place to accept more data, which
would allow us to draw more conclusions about SVMs and limit these effects.
Player
ID for both pitchers and batters proved to be by far the easiest classification
problems to solve, with less than 1% error for the 1-NN classifier in 10-fold
cross-validation, using as few as 5 features. Figure 5 shows why this is the case, namely
that the points for individual players appear to be separated into
player-specific clusters. As is detailed below, this is because the feature
space points appear to be more homogeneously clustered by player than by
motion. As far as methods go, it is
apparent that the performance of k-NN and SVM consistently exceed that of LDA
and NB. The LDA classifier was ineffective because there were often multiple
clusters representing each class that were interspersed in feature space,
making it impossible for a linear discriminant to separate them. The weak performance of Naive Bayes was
likely due to the fact that the distributions of different features were not
independent, as is to be expected given the mechanical relationships among the
various extracted features.
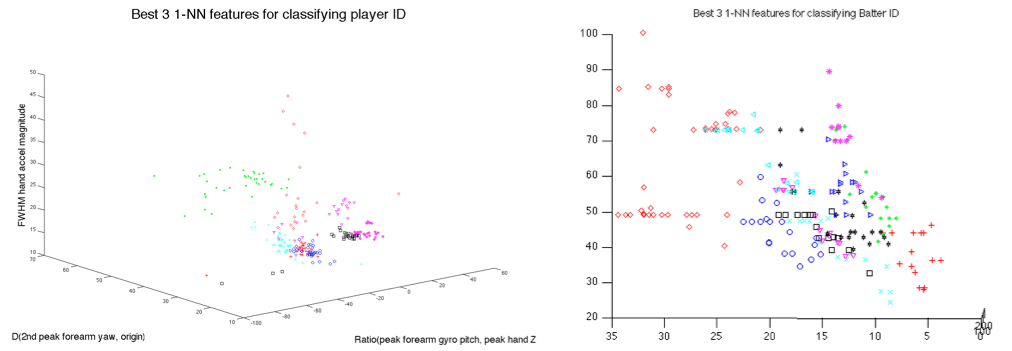
Figure
5: Player-specific clusters are clearly separable for both pitchers and batters
Figure
6 shows all the examples plotted in the 2-d space of the best features selected
for 1-NN classification of pitch type, the relative timing of peaks in hand
gyroscope yaw and hand gyroscope pitch. The figure indicates that the largest
class in the dataset, fastball, is separated in feature space by another class,
breaking balls, suggesting that SVMs should perform better than the other
classifiers. As noted above, feature selection for the binary classifier SVMs
was a challenge due to overfitting on the smallest classes. As a result, to further
explore the potential of SVMs without exposing the method to the same level of
risk of overfitting, we trained SVMs on the top 20 features selected for 5-NN
classification, hoping that features selected by a classifier considering the
entire space of classes might be less prone to overfitting. Note that these
results should not be directly compared with the results above because the SVM
method used different feature sets for each of the binary classification
problems it examined.
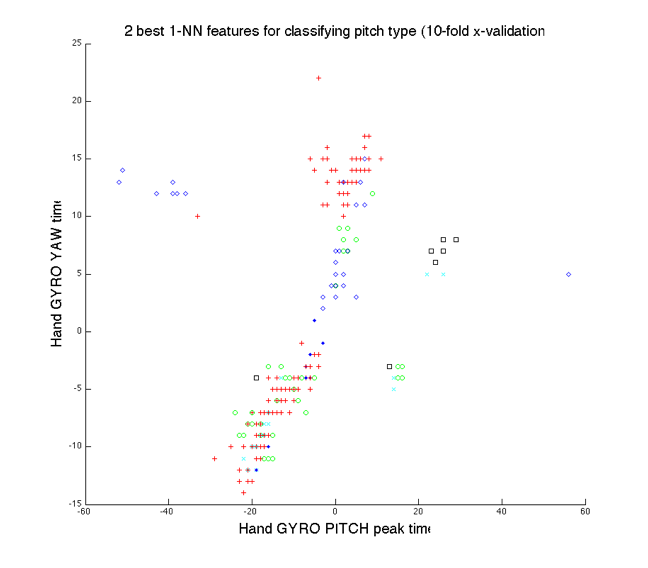
Figure
6: Fastball class is clearly separated by breaking ball class
Using
the k-NN feature set, SVMs out-performed the other classifiers we tried, without
having run feature selection specific to that classifier. We trained SVMs to
classify pitch type using the features selected by the SFS for a 5-NN
classifier, and applied leave-one-out cross-validation to test their
performance. The performance far exceeds that of a naive classifier, especially
for the smallest classes, and would almost certainly benefit from more data for
those classes. Table 5 shows the performance compared to the distribution of
examples. Though we cannot conclude much from this limited test, we believe the
results would improve for the smaller classes if we had more data to describe
them (training and testing with fewer than 10 examples per class is not ideal).
We used the publicly available LibSVM to conduct
these tests [3].
|
Class (distribution
of examples) |
SVM
Performance (true
positives) |
|
1. Fastball
(57% of set) |
85% |
|
2. Change: (17% of set) |
64% |
|
3. Slider: (4% of set) |
45% |
|
4. 2-seam: (7% of set) |
30% |
|
5. 4-seam: (3% of set) |
58% |
|
6. Break: (12% of set) |
70% |
Table
5: SVM compared to prior
distribution of classes.
Varying
the cost parameter for the SVM, which varies the softness of the separating
hyperplane, we found that a lower cost would result in better performance on
the largest class (fastballs), especially as it relates to the rate of false
positives, which increases with fuzzier separation.
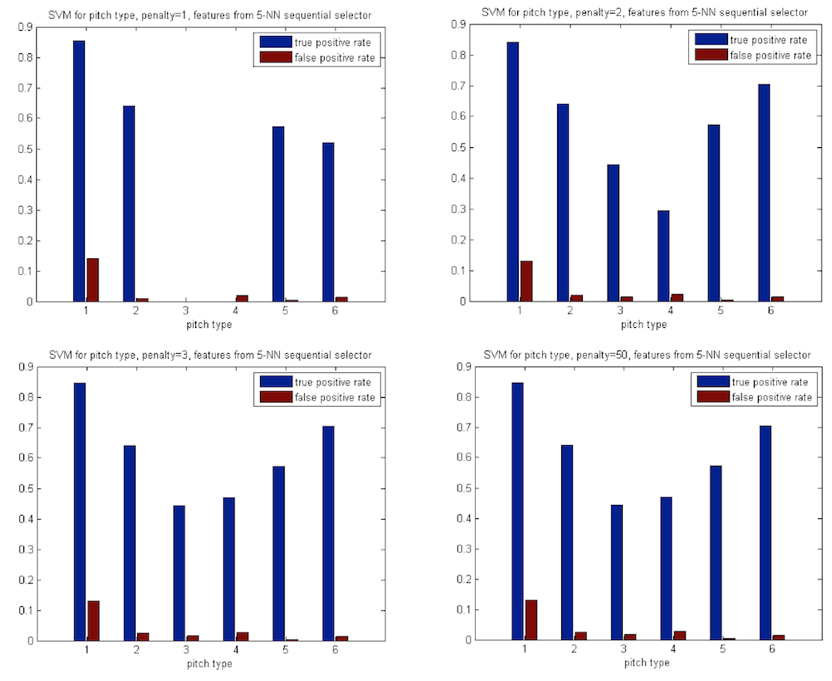
Figure
7: Lower SVM cost parameter results in better performance on the larger class,
worse performance for others.
Conclusion
The
relative performance of the different classifiers indicate a feature space in
which the variability between players is larger than the variability between
motion types. We believe that the performance of k-NN and SVM and the complete
failure of LDA and NB is due to the arrangement of the feature space, in which
the large separation due to subject variability fractures the distributions of
different motion types. K-NN and SVM are far more invariant to this kind of
arrangement than linear and Naive Bayes classifiers, which struggle to linearly
separate the classes. We theorize that data from players with similar
techniques (that result in close proximity in the feature space) would be
easier to classify based on motion type. Indeed, the clear division of the
fastball class into two distinct large clusters (as shown in figure 6) may be
due to a dichotomy in subject technique. With more data, similar player types
could be identified and investigated in separate groups.
Summary
We
applied a set of pattern recognition algorithms to a new dataset of
professional baseball players performing activities such as pitching and
batting in an attempt to classify common techniques for those activities. We
found that these data almost perfectly separate individual players, but
struggle to distinguish amongst player activities. We suggest ruling out LDA
and NB, and focusing on those classification techniques that can better handle
fragmented class distributions.
Future
work
There are many ways
that the study presented here could be improved and extended. The first and obvious way to improve the
results would be to obtain a larger data set, particularly since there are
currently only a few examples of several of the classes. A larger data set
would allow us to more reliably train our classifiers. The data itself may also be improved
through the use of more careful methods for classifying pitch type, as there
are believed to be some misclassified pitches in the database.
More sophisticated
feature selection means could also be implemented. The current SFS method of selecting
features could be improved upon by implementing a Sequential Forward Floating
Search algorithm (SFFS). Such
methods are not “greedy” in that they can improve performance by backtracking,
thereby reconsidering features that could have been erroneously added or
removed. One could also look at
implementing backwards feature selection algorithms (SBS, SBFS), where the
classification performance is evaluated while features are sequentially removed
from the full feature set rather than added to an initially empty feature set.
One drawback of backwards algorithms is that they are typically more
computationally expensive than their forward counterparts. This could be alleviated by using a
forward algorithm to first eliminate some features that are clearly unimportant
and then implementing the backwards algorithm on the remainder. In all these classification methods, it
may be useful to try a “Leave One
Player Out” means of dividing the data set, given the strong player-based clustering
observed here.
In this work, we
began looking at boosting the linear classifiers, without much success, though
further investigation is warranted. Separately, given the results we obtained, a
Decision Tree classification scheme may be worth considering. In this approach,
grouping of players with similar characteristics could represent the first
division. In general, cascading linear classifiers could avoid some of the pitfalls
we encountered using LDA.
Additional
classification algorithms, particularly temporal methods, could potentially
shed more light on this data set.
Temporal methods could be implemented by windowing each data series and
looking at how features change throughout a motion. This could be used to train a Hidden
Markov Model (HMM), for example, which progresses through hidden states as time
evolves. It would be particularly
interesting if such a model was found to correspond to states that coincide
with the accepted biomechanical phases of pitching and hitting.
Finally, there are
many more questions that can be asked using this data, and many other ways that
it could be applied. As seen above,
individual players tend to cluster strongly in some feature space. This observation could be used to
explore player similarities and potentially determine features that identify
different groups of players (those that are successful, those that are injury
prone in a particular way, etc). Another class label that could be
explored is player fatigue, which would be indexed simply by the timing of a
gesture in a data session. One
could look at the changes that occur in features as a player tires and classify
fatigue levels based on this information.
This has obvious sports medicine implications, as it could help explore
potential sources of injury as a player tires.

Figure
7: Fortune cookie (from 12/9/10) confirms that 42.7% of all statistics are made
up on the spot.
Code
Download
our Matlab code at:
http://www.media.mit.edu/~gershon/MAS622j/final_project/code.zip
References:
[1]
Michael Lapinski , A Wearable, Wireless Sensor System
for Sports Medicine, MS Thesis, MIT Media Lab, September 2008
[2]
Richard O. Duda, Peter E. Hart, and David G. Stork,
Pattern Classification. New York, NY: John Wiley & Sons
[3]
Chih-Chung Chang and Chih-Jen
Lin, LIBSVM : a library for support vector machines, 2001. Software available
at http://www.csie.ntu.edu.tw/~cjlin/libsvm